This is a web interface of the Probe Particle Model for simulation of high-resolution atomic force microscopy images (see references to publications below).
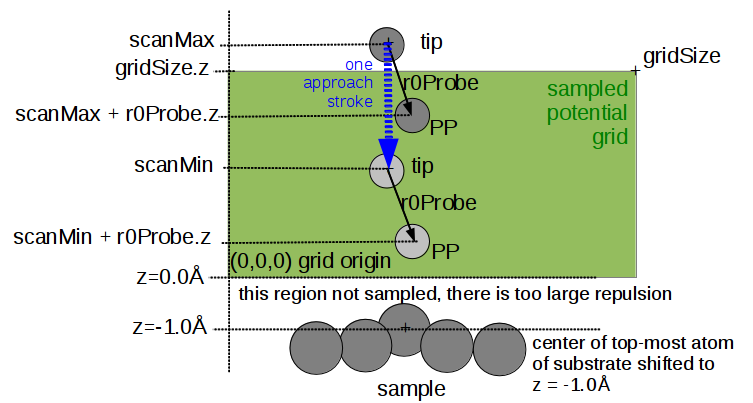
The geometry of the structure supplied in .xyz format is automatically shifted so that the topmost atom of sample is positioned -1.0Å below zero (zero means closest approach of probe particle). Closer approaches would lead to unrealisticaly high repulsive forces.
The geometry alignment also depends on setting of the periodic boundary conditions (PBC) mode. For a PBC = True setting, the sampling grid is defined manually by setting gridA, gridB, gridC vectors. For non-periodic boundary conditions (PBC = False) an orthogonal sampling grid around the molecule is created automatically with margins 3.0Å on each side.
In this web version of the code, it is possible to include electrostatic field of sample approximated only by partial atomic charges (i.e. point charges). To do so, just put appropriated partial atomic charge [e+] into the 5th column of your input geometry (the .xyz file). For lines where 5th column is missing, the partial atomic charge Qi is automatically assumed to be zero.
WARNING: In case of periodic systems (PBC = True), please ensure that the sum of all charges of all atoms in the system is zero. If there is nonzero net charge of the supercell, the electrostatic force decays very slowly. This would lead to artefacts on the borders of the sampling grid, since we consider interaction just to the neares neighbor supercell.
NOTE: For more accurate description of the electrostatics, it is possible to download the full code from the links bellow, which allows to read the electrostatic forcefield on a grid (e.g. from an XCrysDen .xsf file).
The full code can be downloaded from github
here. There are two implementations available:
a) Python + C++ implementation used on this webpage
or
b) older Fortran90 implementation used in the publications cited below.
For more detailed information including technical details, please, see our github wiki
In case you use the Probe Particle Model for your research, consider citing this papers:
Prokop Hapala, Georgy Kichin, Christian Wagner, F. Stefan Tautz, Ruslan Temirov, and Pavel Jelínek, Mechanism of high-resolution STM/AFM imaging with functionalized tips, Phys. Rev. B 90, 085421 – Published 19 August 2014
Prokop Hapala, Ruslan Temirov, F. Stefan Tautz, and Pavel Jelínek, Origin of High-Resolution IETS-STM Images of Organic Molecules with Functionalized Tips, Phys. Rev. Lett. 113, 226101 – Published 25 November 2014